Program
Here is another example of a complete program that demonstrates several of the key functions in the libr package. The example also shows how the package integrates with sassy.
library(sassy)
options("logr.autolog" = TRUE,
"logr.notes" = FALSE)
# Get temp location for log and report output
tmp <- tempdir()
# Open log
lf <- log_open(file.path(tmp, "example1.log"))
# Prepare Data ------------------------------------------------------------
sep("Prepare Data")
# Get path to sample data
pkg <- system.file("extdata", package = "libr")
# Create libname for csv data
libname(sdtm, pkg, "csv", quiet = TRUE)
put("Join and prepare data")
put("Join DM to VS and keep desired columns")
datastep(sdtm$DM, merge = sdtm$VS, merge_by = USUBJID,
keep = v(USUBJID, VSTESTCD, VISIT, VISITNUM, VSSTRESN, ARM, VSBLFL),
where = expression(VSTESTCD %in% c("PULSE", "RESP", "TEMP", "DIABP", "SYSBP") &
!(VISIT == "SCREENING" & VSBLFL != "Y")), {}) -> dm_joined
put("Sort by variables")
proc_sort(dm_joined, by = v(USUBJID, VSTESTCD, VISITNUM)) -> dm_sorted
put("Differentiate baseline from treated vital signs")
datastep(dm_sorted, by = v(USUBJID, VSTESTCD),
retain = list(BSTRESN = 0), {
# Combine treatment groups
# And distinguish baseline time points
if (ARM == "ARM A") {
if (VSBLFL %eq% "Y") {
GRP <- "A_BASE"
} else {
GRP <- "A_TRT"
}
} else {
if (VSBLFL %eq% "Y") {
GRP <- "O_BASE"
} else {
GRP <- "O_TRT"
}
}
# Populate baseline value
if (first.)
BSTRESN = VSSTRESN
}) -> prep
put("Get population counts")
pop_A <- subset(prep, GRP == "A_BASE", v(USUBJID, GRP)) |>
proc_sort(options = nodupkey) |>
proc_freq(tables = GRP,
options = v(nocum, nonobs, nopercent),
output = long) |>
subset(select = "A_BASE", drop = TRUE)
pop_O <- subset(prep, GRP == "O_BASE", v(USUBJID, GRP)) |>
proc_sort(options = nodupkey) |>
proc_freq(tables = GRP,
options = v(nocum, nonobs, nopercent),
output = long) |>
subset(select = "O_BASE", drop = TRUE)
# Prepare formats ---------------------------------------------------------
sep("Prepare formats")
put("Vital sign lookup format")
vs_fmt <- c(PULSE = "Pulse",
TEMP = "Temperature °C",
RESP = "Respirations/min",
SYSBP = "Systolic Blood Pressure",
DIABP = "Diastolic Blood Pressure") |> put()
put("Statistics lookup format")
stat_fmt <- c(MEANSTD = "Mean (SD)",
MEDIAN = "Median",
Q1Q3 = "Q1 - Q3",
MINMAX = "Min - Max") |> put()
put("Create format catalog")
fc <- fcat(MEAN = "%.1f",
STD = "(%.2f)",
MEDIAN = "%.1f",
Q1 = "%.1f",
Q3 = "%.1f",
MIN = "%.1f",
MAX = "%.1f")
# Prepare final data frame ------------------------------------------------
sep("Prepare final data")
put("Calculate statistics and prepare final data frame")
proc_means(prep, var = VSSTRESN, class = VSTESTCD, by = GRP,
stats = v(mean, std, median, q1, q3, min, max),
options = v(notype, nofreq, nway)) |>
datastep(format = fc,
drop = v(MEAN, STD, Q1, Q3, MIN, MAX, VAR),
rename = c("CLASS" = "VAR"),
{
MEANSTD <- fapply2(MEAN, STD)
Q1Q3 <- fapply2(Q1, Q3, sep = " - ")
MINMAX <- fapply2(MIN, MAX, sep = " - ")
}) |>
proc_transpose(id = BY, var = v(MEANSTD, MEDIAN, Q1Q3, MINMAX),
by = VAR, name = "LABEL") -> final
put("Prepare factor for sorting")
final$VAR <- factor(final$VAR, names(vs_fmt))
put("Final sort")
proc_sort(final, by = VAR) -> final
# Create Report -----------------------------------------------------------
sep("Create Report")
# Define table object
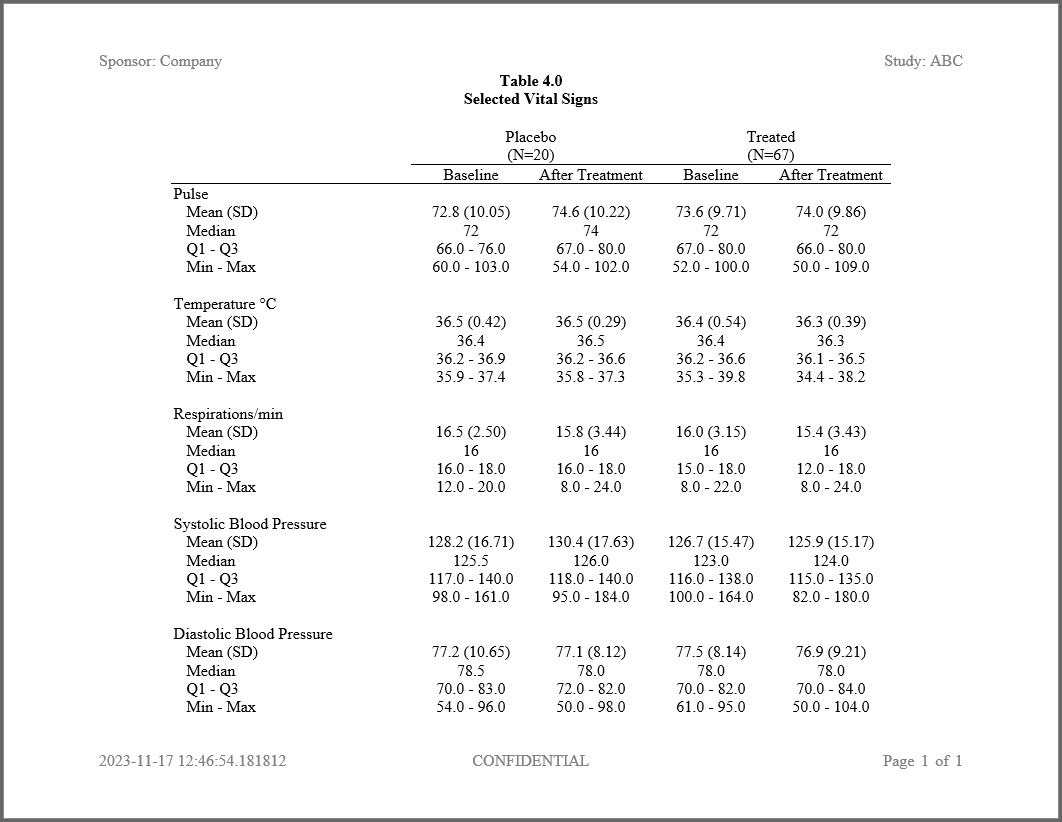
tbl <- create_table(final) |>
spanning_header(A_BASE, A_TRT, "Placebo", n = pop_A) |>
spanning_header(O_BASE, O_TRT, "Treated", n = pop_O) |>
column_defaults(width = 1.25, align = "center") |>
stub(c(VAR, LABEL), width = 2.5) |>
define(VAR, "Vital Sign", format = vs_fmt,
blank_after = TRUE, dedupe = TRUE, label_row = TRUE) |>
define(LABEL, indent = .25, format = stat_fmt) |>
define(A_BASE, "Baseline") |>
define(A_TRT, "After Treatment") |>
define(O_BASE, "Baseline") |>
define(O_TRT, "After Treatment")
# Define report object
rpt <- create_report(file.path(tmp, "./output/example2.rtf"), output_type = "RTF",
font = "Times", font_size = 12) |>
page_header("Sponsor: Company", "Study: ABC") |>
titles("Table 4.0", "Selected Vital Signs", bold = TRUE) |>
add_content(tbl, align = "center") |>
page_footer(Sys.time(), "CONFIDENTIAL", "Page [pg] of [tpg]")
# Write report to file system
res <- write_report(rpt)
# Clean Up ----------------------------------------------------------------
sep("Clean Up")
# Close log
log_close()
# View report
# file.show(res$file_path)
# View log
# file.show(lf)Log
Here is the log from the above program:
=========================================================================
Log Path: C:/Users/dbosa/AppData/Local/Temp/RtmpiipZo8/log/example1.log
Program Path: C:/packages/Testing/Example2.R
Working Directory: C:/packages/Testing
User Name: dbosa
R Version: 4.3.1 (2023-06-16 ucrt)
Machine: SOCRATES x86-64
Operating System: Windows 10 x64 build 22621
Base Packages: stats graphics grDevices utils datasets methods base Other
Packages: tidylog_1.0.2 procs_1.0.3 reporter_1.4.2 libr_1.2.8 logr_1.3.5
fmtr_1.6.1 common_1.1.0 sassy_1.2.1
Log Start Time: 2023-11-17 13:03:47.835721
=========================================================================
=========================================================================
Prepare Data
=========================================================================
# library 'sdtm': 8 items
- attributes: csv not loaded
- path: C:/Users/dbosa/AppData/Local/R/win-library/4.3/libr/extdata
- items:
Name Extension Rows Cols Size LastModified
1 AE csv 150 27 88.5 Kb 2023-09-09 22:45:51
2 DA csv 3587 18 528.2 Kb 2023-09-09 22:45:51
3 DM csv 87 24 45.5 Kb 2023-09-09 22:45:51
4 DS csv 174 9 34.1 Kb 2023-09-09 22:45:51
5 EX csv 84 11 26.4 Kb 2023-09-09 22:45:51
6 IE csv 2 14 13.4 Kb 2023-09-09 22:45:51
7 SV csv 685 10 70.3 Kb 2023-09-09 22:45:51
8 VS csv 3358 17 467.4 Kb 2023-09-09 22:45:51
Join and prepare data
Join DM to VS and keep desired columns
datastep: columns decreased from 24 to 7
# A tibble: 2,768 × 7
USUBJID VSTESTCD VISIT VISITNUM VSSTRESN ARM VSBLFL
<chr> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 ABC-01-049 DIABP DAY 1 1 76 ARM D Y
2 ABC-01-049 DIABP WEEK 2 2 66 ARM D <NA>
3 ABC-01-049 DIABP WEEK 4 4 84 ARM D <NA>
4 ABC-01-049 DIABP WEEK 6 6 68 ARM D <NA>
5 ABC-01-049 DIABP WEEK 8 8 80 ARM D <NA>
6 ABC-01-049 DIABP WEEK 12 12 70 ARM D <NA>
7 ABC-01-049 DIABP WEEK 16 16 70 ARM D <NA>
8 ABC-01-049 PULSE DAY 1 1 84 ARM D Y
9 ABC-01-049 PULSE WEEK 2 2 84 ARM D <NA>
10 ABC-01-049 PULSE WEEK 4 4 76 ARM D <NA>
# ℹ 2,758 more rows
# ℹ Use `print(n = ...)` to see more rows
Sort by variables
proc_sort: input data set 2768 rows and 7 columns
by: USUBJID VSTESTCD VISITNUM
keep: USUBJID VSTESTCD VISIT VISITNUM VSSTRESN ARM VSBLFL
order: a a a
output data set 2768 rows and 7 columns
# A tibble: 2,768 × 7
USUBJID VSTESTCD VISIT VISITNUM VSSTRESN ARM VSBLFL
<chr> <chr> <chr> <dbl> <dbl> <chr> <chr>
1 ABC-01-049 DIABP DAY 1 1 76 ARM D Y
2 ABC-01-049 DIABP WEEK 2 2 66 ARM D <NA>
3 ABC-01-049 DIABP WEEK 4 4 84 ARM D <NA>
4 ABC-01-049 DIABP WEEK 6 6 68 ARM D <NA>
5 ABC-01-049 DIABP WEEK 8 8 80 ARM D <NA>
6 ABC-01-049 DIABP WEEK 12 12 70 ARM D <NA>
7 ABC-01-049 DIABP WEEK 16 16 70 ARM D <NA>
8 ABC-01-049 PULSE DAY 1 1 84 ARM D Y
9 ABC-01-049 PULSE WEEK 2 2 84 ARM D <NA>
10 ABC-01-049 PULSE WEEK 4 4 76 ARM D <NA>
# ℹ 2,758 more rows
# ℹ Use `print(n = ...)` to see more rows
Differentiate baseline from treated vital signs
datastep: columns increased from 7 to 9
# A tibble: 2,768 × 9
USUBJID VSTESTCD VISIT VISITNUM VSSTRESN ARM VSBLFL GRP BSTRESN
<chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <dbl>
1 ABC-01-049 DIABP DAY 1 1 76 ARM D Y O_BASE 76
2 ABC-01-049 DIABP WEEK 2 2 66 ARM D <NA> O_TRT 76
3 ABC-01-049 DIABP WEEK 4 4 84 ARM D <NA> O_TRT 76
4 ABC-01-049 DIABP WEEK 6 6 68 ARM D <NA> O_TRT 76
5 ABC-01-049 DIABP WEEK 8 8 80 ARM D <NA> O_TRT 76
6 ABC-01-049 DIABP WEEK 12 12 70 ARM D <NA> O_TRT 76
7 ABC-01-049 DIABP WEEK 16 16 70 ARM D <NA> O_TRT 76
8 ABC-01-049 PULSE DAY 1 1 84 ARM D Y O_BASE 84
9 ABC-01-049 PULSE WEEK 2 2 84 ARM D <NA> O_TRT 84
10 ABC-01-049 PULSE WEEK 4 4 76 ARM D <NA> O_TRT 84
# ℹ 2,758 more rows
# ℹ Use `print(n = ...)` to see more rows
Get population counts
proc_sort: input data set 20 rows and 2 columns
by: USUBJID GRP
keep: USUBJID GRP
order: a
options: nodupkey
output data set 20 rows and 2 columns
# A tibble: 20 × 2
USUBJID GRP
<chr> <chr>
1 ABC-01-051 A_BASE
2 ABC-01-056 A_BASE
3 ABC-02-034 A_BASE
4 ABC-02-038 A_BASE
5 ABC-02-109 A_BASE
6 ABC-03-002 A_BASE
7 ABC-03-006 A_BASE
8 ABC-03-091 A_BASE
9 ABC-04-075 A_BASE
10 ABC-04-080 A_BASE
11 ABC-04-126 A_BASE
12 ABC-06-068 A_BASE
13 ABC-06-069 A_BASE
14 ABC-07-012 A_BASE
15 ABC-07-016 A_BASE
16 ABC-08-104 A_BASE
17 ABC-08-106 A_BASE
18 ABC-09-020 A_BASE
19 ABC-09-023 A_BASE
20 ABC-09-137 A_BASE
proc_freq: input data set 20 rows and 2 columns
tables: GRP
output: long
view: TRUE
output: 1 datasets
# A tibble: 1 × 3
VAR STAT A_BASE
<chr> <chr> <dbl>
1 GRP CNT 20
proc_sort: input data set 67 rows and 2 columns
by: USUBJID GRP
keep: USUBJID GRP
order: a
options: nodupkey
output data set 67 rows and 2 columns
# A tibble: 67 × 2
USUBJID GRP
<chr> <chr>
1 ABC-01-049 O_BASE
2 ABC-01-050 O_BASE
3 ABC-01-052 O_BASE
4 ABC-01-053 O_BASE
5 ABC-01-054 O_BASE
6 ABC-01-055 O_BASE
7 ABC-01-113 O_BASE
8 ABC-01-114 O_BASE
9 ABC-02-033 O_BASE
10 ABC-02-035 O_BASE
# ℹ 57 more rows
# ℹ Use `print(n = ...)` to see more rows
proc_freq: input data set 67 rows and 2 columns
tables: GRP
output: long
view: TRUE
output: 1 datasets
# A tibble: 1 × 3
VAR STAT O_BASE
<chr> <chr> <dbl>
1 GRP CNT 67
=========================================================================
Prepare formats
=========================================================================
Vital sign lookup format
Pulse
Temperature °C
Respirations/min
Systolic Blood Pressure
Diastolic Blood Pressure
Statistics lookup format
Mean (SD)
Median
Q1 - Q3
Min - Max
Create format catalog
# A format catalog: 7 formats
- $MEAN: type S, "%.1f"
- $STD: type S, "(%.2f)"
- $MEDIAN: type S, "%.1f"
- $Q1: type S, "%.1f"
- $Q3: type S, "%.1f"
- $MIN: type S, "%.1f"
- $MAX: type S, "%.1f"
=========================================================================
Prepare final data
=========================================================================
Calculate statistics and prepare final data frame
proc_means: input data set 2768 rows and 9 columns
by: GRP
class: VSTESTCD
var: VSSTRESN
stats: mean std median q1 q3 min max
view: TRUE
output: 1 datasets
CLASS BY VAR MEAN STD MEDIAN Q1 Q3 MIN MAX
1 DIABP A_BASE VSSTRESN 77.15000 10.6537614 78.5 70.0 83.0 54.0 96.0
2 PULSE A_BASE VSSTRESN 72.75000 10.0518393 72.0 66.0 76.0 60.0 103.0
3 RESP A_BASE VSSTRESN 16.50000 2.5026302 16.0 16.0 18.0 12.0 20.0
4 SYSBP A_BASE VSSTRESN 128.15000 16.7120347 125.5 117.0 140.0 98.0 161.0
5 TEMP A_BASE VSSTRESN 36.52105 0.4171050 36.4 36.2 36.9 35.9 37.4
6 DIABP A_TRT VSSTRESN 77.08547 8.1182772 78.0 72.0 82.0 50.0 98.0
7 PULSE A_TRT VSSTRESN 74.58120 10.2201958 74.0 67.0 80.0 54.0 102.0
8 RESP A_TRT VSSTRESN 15.80342 3.4372185 16.0 16.0 18.0 8.0 24.0
9 SYSBP A_TRT VSSTRESN 130.43590 17.6304911 126.0 118.0 140.0 95.0 184.0
10 TEMP A_TRT VSSTRESN 36.45431 0.2857157 36.5 36.2 36.6 35.8 37.3
11 DIABP O_BASE VSSTRESN 77.48485 8.1415221 78.0 70.0 82.0 61.0 95.0
12 PULSE O_BASE VSSTRESN 73.55224 9.7051745 72.0 67.0 80.0 52.0 100.0
13 RESP O_BASE VSSTRESN 15.97015 3.1477272 16.0 15.0 18.0 8.0 22.0
14 SYSBP O_BASE VSSTRESN 126.68182 15.4672449 123.0 116.0 138.0 100.0 164.0
15 TEMP O_BASE VSSTRESN 36.42239 0.5376213 36.4 36.2 36.6 35.3 39.8
16 DIABP O_TRT VSSTRESN 76.94286 9.2058374 78.0 70.0 84.0 50.0 104.0
17 PULSE O_TRT VSSTRESN 74.02279 9.8592694 72.0 66.0 80.0 50.0 109.0
18 RESP O_TRT VSSTRESN 15.44444 3.4312291 16.0 12.0 18.0 8.0 24.0
19 SYSBP O_TRT VSSTRESN 125.90571 15.1715962 124.0 115.0 135.0 82.0 180.0
20 TEMP O_TRT VSSTRESN 36.31057 0.3903231 36.3 36.1 36.5 34.4 38.2
datastep: columns decreased from 10 to 6
VAR BY MEDIAN MEANSTD Q1Q3 MINMAX
1 DIABP A_BASE 78.5 77.2 (10.65) 70.0 - 83.0 54.0 - 96.0
2 PULSE A_BASE 72.0 72.8 (10.05) 66.0 - 76.0 60.0 - 103.0
3 RESP A_BASE 16.0 16.5 (2.50) 16.0 - 18.0 12.0 - 20.0
4 SYSBP A_BASE 125.5 128.2 (16.71) 117.0 - 140.0 98.0 - 161.0
5 TEMP A_BASE 36.4 36.5 (0.42) 36.2 - 36.9 35.9 - 37.4
6 DIABP A_TRT 78.0 77.1 (8.12) 72.0 - 82.0 50.0 - 98.0
7 PULSE A_TRT 74.0 74.6 (10.22) 67.0 - 80.0 54.0 - 102.0
8 RESP A_TRT 16.0 15.8 (3.44) 16.0 - 18.0 8.0 - 24.0
9 SYSBP A_TRT 126.0 130.4 (17.63) 118.0 - 140.0 95.0 - 184.0
10 TEMP A_TRT 36.5 36.5 (0.29) 36.2 - 36.6 35.8 - 37.3
11 DIABP O_BASE 78.0 77.5 (8.14) 70.0 - 82.0 61.0 - 95.0
12 PULSE O_BASE 72.0 73.6 (9.71) 67.0 - 80.0 52.0 - 100.0
13 RESP O_BASE 16.0 16.0 (3.15) 15.0 - 18.0 8.0 - 22.0
14 SYSBP O_BASE 123.0 126.7 (15.47) 116.0 - 138.0 100.0 - 164.0
15 TEMP O_BASE 36.4 36.4 (0.54) 36.2 - 36.6 35.3 - 39.8
16 DIABP O_TRT 78.0 76.9 (9.21) 70.0 - 84.0 50.0 - 104.0
17 PULSE O_TRT 72.0 74.0 (9.86) 66.0 - 80.0 50.0 - 109.0
18 RESP O_TRT 16.0 15.4 (3.43) 12.0 - 18.0 8.0 - 24.0
19 SYSBP O_TRT 124.0 125.9 (15.17) 115.0 - 135.0 82.0 - 180.0
20 TEMP O_TRT 36.3 36.3 (0.39) 36.1 - 36.5 34.4 - 38.2
proc_transpose: input data set 20 rows and 6 columns
by: VAR
var: MEANSTD MEDIAN Q1Q3 MINMAX
id: BY
name: LABEL
output dataset 20 rows and 6 columns
VAR LABEL A_BASE A_TRT O_BASE O_TRT
1 DIABP MEANSTD 77.2 (10.65) 77.1 (8.12) 77.5 (8.14) 76.9 (9.21)
2 DIABP MEDIAN 78.5 78.0 78.0 78.0
3 DIABP Q1Q3 70.0 - 83.0 72.0 - 82.0 70.0 - 82.0 70.0 - 84.0
4 DIABP MINMAX 54.0 - 96.0 50.0 - 98.0 61.0 - 95.0 50.0 - 104.0
5 PULSE MEANSTD 72.8 (10.05) 74.6 (10.22) 73.6 (9.71) 74.0 (9.86)
6 PULSE MEDIAN 72 74 72 72
7 PULSE Q1Q3 66.0 - 76.0 67.0 - 80.0 67.0 - 80.0 66.0 - 80.0
8 PULSE MINMAX 60.0 - 103.0 54.0 - 102.0 52.0 - 100.0 50.0 - 109.0
9 RESP MEANSTD 16.5 (2.50) 15.8 (3.44) 16.0 (3.15) 15.4 (3.43)
10 RESP MEDIAN 16 16 16 16
11 RESP Q1Q3 16.0 - 18.0 16.0 - 18.0 15.0 - 18.0 12.0 - 18.0
12 RESP MINMAX 12.0 - 20.0 8.0 - 24.0 8.0 - 22.0 8.0 - 24.0
13 SYSBP MEANSTD 128.2 (16.71) 130.4 (17.63) 126.7 (15.47) 125.9 (15.17)
14 SYSBP MEDIAN 125.5 126.0 123.0 124.0
15 SYSBP Q1Q3 117.0 - 140.0 118.0 - 140.0 116.0 - 138.0 115.0 - 135.0
16 SYSBP MINMAX 98.0 - 161.0 95.0 - 184.0 100.0 - 164.0 82.0 - 180.0
17 TEMP MEANSTD 36.5 (0.42) 36.5 (0.29) 36.4 (0.54) 36.3 (0.39)
18 TEMP MEDIAN 36.4 36.5 36.4 36.3
19 TEMP Q1Q3 36.2 - 36.9 36.2 - 36.6 36.2 - 36.6 36.1 - 36.5
20 TEMP MINMAX 35.9 - 37.4 35.8 - 37.3 35.3 - 39.8 34.4 - 38.2
Prepare factor for sorting
Final sort
proc_sort: input data set 20 rows and 6 columns
by: VAR
keep: VAR LABEL A_BASE A_TRT O_BASE O_TRT
order: a
output data set 20 rows and 6 columns
VAR LABEL A_BASE A_TRT O_BASE O_TRT
5 PULSE MEANSTD 72.8 (10.05) 74.6 (10.22) 73.6 (9.71) 74.0 (9.86)
6 PULSE MEDIAN 72 74 72 72
7 PULSE Q1Q3 66.0 - 76.0 67.0 - 80.0 67.0 - 80.0 66.0 - 80.0
8 PULSE MINMAX 60.0 - 103.0 54.0 - 102.0 52.0 - 100.0 50.0 - 109.0
17 TEMP MEANSTD 36.5 (0.42) 36.5 (0.29) 36.4 (0.54) 36.3 (0.39)
18 TEMP MEDIAN 36.4 36.5 36.4 36.3
19 TEMP Q1Q3 36.2 - 36.9 36.2 - 36.6 36.2 - 36.6 36.1 - 36.5
20 TEMP MINMAX 35.9 - 37.4 35.8 - 37.3 35.3 - 39.8 34.4 - 38.2
9 RESP MEANSTD 16.5 (2.50) 15.8 (3.44) 16.0 (3.15) 15.4 (3.43)
10 RESP MEDIAN 16 16 16 16
11 RESP Q1Q3 16.0 - 18.0 16.0 - 18.0 15.0 - 18.0 12.0 - 18.0
12 RESP MINMAX 12.0 - 20.0 8.0 - 24.0 8.0 - 22.0 8.0 - 24.0
13 SYSBP MEANSTD 128.2 (16.71) 130.4 (17.63) 126.7 (15.47) 125.9 (15.17)
14 SYSBP MEDIAN 125.5 126.0 123.0 124.0
15 SYSBP Q1Q3 117.0 - 140.0 118.0 - 140.0 116.0 - 138.0 115.0 - 135.0
16 SYSBP MINMAX 98.0 - 161.0 95.0 - 184.0 100.0 - 164.0 82.0 - 180.0
1 DIABP MEANSTD 77.2 (10.65) 77.1 (8.12) 77.5 (8.14) 76.9 (9.21)
2 DIABP MEDIAN 78.5 78.0 78.0 78.0
3 DIABP Q1Q3 70.0 - 83.0 72.0 - 82.0 70.0 - 82.0 70.0 - 84.0
4 DIABP MINMAX 54.0 - 96.0 50.0 - 98.0 61.0 - 95.0 50.0 - 104.0
=========================================================================
Create Report
=========================================================================
# A report specification: 1 pages
- file_path: 'C:\Users\dbosa\AppData\Local\Temp\RtmpiipZo8/./output/example2.rtf'
- output_type: RTF
- units: inches
- orientation: landscape
- margins: top 0.5 bottom 0.5 left 1 right 1
- line size/count: 9/38
- page_header: left=Sponsor: Company right=Study: ABC
- title 1: 'Table 4.0'
- title 2: 'Selected Vital Signs'
- page_footer: left=2023-11-17 13:03:53.809731 center=CONFIDENTIAL right=Page [pg] of [tpg]
- content:
# A table specification:
- data: data.frame 'final' 20 rows 6 cols
- show_cols: all
- use_attributes: all
- spanning_header: from='A_BASE' to='A_TRT' 'Placebo' level=1
- spanning_header: from='O_BASE' to='O_TRT' 'Treated' level=1
- stub: VAR LABEL width=2.5 align='left'
- define: VAR 'Vital Sign' dedupe='TRUE'
- define: LABEL
- define: A_BASE 'Baseline'
- define: A_TRT 'After Treatment'
- define: O_BASE 'Baseline'
- define: O_TRT 'After Treatment'
=========================================================================
Clean Up
=========================================================================
=========================================================================
Log End Time: 2023-11-17 13:03:54.012811
Log Elapsed Time: 0 00:00:06
=========================================================================